 W
W3D Slicer (Slicer) is a free and open source software package for image analysis and scientific visualization. Slicer is used in a variety of medical applications, including autism, multiple sclerosis, systemic lupus erythematosus, prostate cancer, lung cancer, breast cancer, schizophrenia, orthopedic biomechanics, COPD, cardiovascular disease and neurosurgery.
 W
WThe Aphelion Imaging Software Suite is a software suite that includes three base products - Aphelion Lab, Aphelion Dev, and Aphelion SDK for addressing image processing and image analysis applications. The suite also includes a set of extension programs to implement specific vertical applications that benefit from imaging techniques.
 W
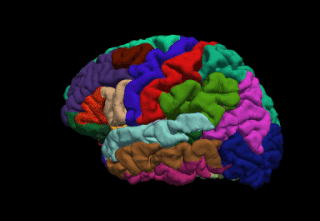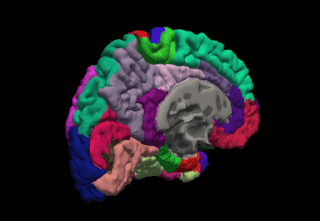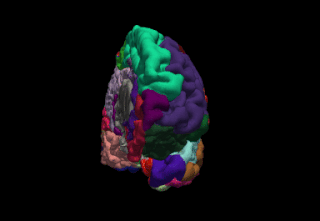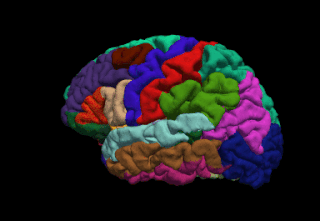
WFreeSurfer is a brain imaging software package originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.
 W
WGinkgo CADx is a multi platform DICOM viewer (*.dcm) and dicomizer. Ginkgo CADx is licensed under LGPL license, being an open source project with an Open core approach. The goal of Ginkgo CADx project is to develop an open source professional DICOM workstation.
 W
WInVesalius is a free medical software used to generate virtual reconstructions of structures in the human body. Based on two-dimensional images, acquired using computed tomography or magnetic resonance imaging equipment, the software generates virtual three-dimensional models correspondent to anatomical parts of the human body. After constructing three-dimensional DICOM images, the software allows the generation of STL (stereolithography) files. These files can be used for rapid prototyping.
 W
WMayaVi is a scientific data visualizer written in Python, which uses VTK and provides a GUI via Tkinter. MayaVi was developed by Prabhu Ramachandran, is free and distributed under the BSD License. It is cross-platform and runs on any platform where both Python and VTK are available. MayaVi is pronounced as a single name, "Ma-ya-vee", meaning "magical" in Sanskrit. The code of MayaVi has nothing in common with that of Autodesk Maya or the Vi text editor.
 W
WMolekel is a free software multiplatform molecular visualization program. It was originally developed at the University of Geneva by Peter F. Flükiger in the 1990s for Silicon Graphics Computers. In 1998, Stefan Portmann took over responsibility and released Version 3.0. Version 4.0 was a nearly platform independent version. Further developments lead to version 4.3, before Stefan Portmann moved on and ceased to develop the codes. In 2006, the Swiss National Supercomputing Centre (CSCS) restarted the project and version 5.0 was released on 21 December 2006.
 W
WMOOSE is an object-oriented C++ finite element framework for the development of tightly coupled multiphysics solvers from Idaho National Laboratory. MOOSE makes use of the PETSc non-linear solver package and libmesh to provide the finite element discretization.
 W
WParaView is an open-source multiple-platform application for interactive, scientific visualization. It has a client–server architecture to facilitate remote visualization of datasets, and generates level of detail (LOD) models to maintain interactive frame rates for large datasets. It is an application built on top of the Visualization Toolkit (VTK) libraries. ParaView is an application designed for data parallelism on shared-memory or distributed-memory multicomputers and clusters. It can also be run as a single-computer application.
 W
WStudierfenster is a free, non-commercial Open Science client/server-based Medical Imaging Processing (MIP) online framework. It offers capabilities, like viewing medical data in two-dimensional (2D) and three-dimensional space (3D) directly in a standard web browser, like Google Chrome, Mozilla Firefox, Safari or Microsoft Edge. Other functionalities are the calculation of Medical Metrics, manual slice-by-slice outlining of structures in medical images (segmentation), manual placing of (anatomical) landmarks in medical image data, viewing medical data in Virtual Reality (VR) and a facial reconstruction and registration of medical data for Augmented Reality (AR).
 W
WVisIt is an open-source interactive parallel visualization and graphical analysis tool for viewing scientific data. It can be used to visualize scalar and vector fields defined on 2D and 3D structured and unstructured meshes. VisIt was designed to handle very large data set sizes in the terascale range and yet can also handle small data sets in the kilobyte range.
 W
WVisTrails is a scientific workflow management system developed at the Scientific Computing and Imaging Institute at the University of Utah that provides support for data exploration and visualization. It is written in Python and employs Qt via PyQt bindings. The system is open source, released under the GPL v2 license. The pre-compiled versions for Windows, Mac OS X, and Linux come with an installer and several packages, including VTK, matplotlib, and ImageMagick. VisTrails also supports user-defined packages.