 W
WDNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
 W
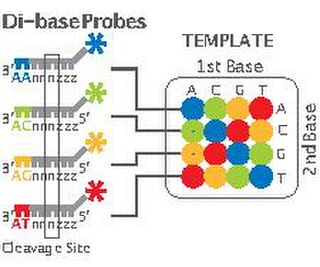
W2 Base Encoding, also called SOLiD, is a next-generation sequencing technology developed by Applied Biosystems and has been commercially available since 2008. These technologies generate hundreds of thousands of small sequence reads at one time. Well-known examples of such DNA sequencing methods include 454 pyrosequencing, the Solexa system and the SOLiD system. These methods have reduced the cost from $0.01/base in 2004 to nearly $0.0001/base in 2006 and increased the sequencing capacity from 1,000,000 bases/machine/day in 2004 to more than 100,000,000 bases/machine/day in 2006.
 W
WSOLiD (Sequencing by Oligonucleotide Ligation and Detection) is a next-generation DNA sequencing technology developed by Life Technologies and has been commercially available since 2006. This next generation technology generates 108 - 109 small sequence reads at one time. It uses 2 base encoding to decode the raw data generated by the sequencing platform into sequence data.
 W
WBGI, currently known as the BGI Group, formerly known as the Beijing Genomics Institute, is a Chinese genome sequencing company, headquartered in Shenzhen, Guangdong, China.
 W
WThe Carlson curve is a term to describe the rate of DNA sequencing or cost per sequenced base as a function of time. It is the biotechnological equivalent of Moore's law. Robert Carlson predicted that the doubling time of DNA sequencing technologies would be at least as fast as Moore's law.
 W
WChIP-exo is a chromatin immunoprecipitation based method for mapping the locations at which a protein of interest binds to the genome. It is a modification of the ChIP-seq protocol, improving the resolution of binding sites from hundreds of base pairs to almost one base pair. It employs the use of exonucleases to degrade strands of the protein-bound DNA in the 5'-3' direction to within a small number of nucleotides of the protein binding site. The nucleotides of the exonuclease-treated ends are determined using some combination of DNA sequencing, microarrays, and PCR. These sequences are then mapped to the genome to identify the locations on the genome at which the protein binds.
 W
WDNA nanoball sequencing is a high throughput sequencing technology that is used to determine the entire genomic sequence of an organism. The method uses rolling circle replication to amplify small fragments of genomic DNA into DNA nanoballs. Fluorescent nucleotides bind to complementary nucleotides and are then polymerized to anchor sequences bound to known sequences on the DNA template. The base order is determined via the fluorescence of the bound nucleotides This DNA sequencing method allows large numbers of DNA nanoballs to be sequenced per run at lower reagent costs compared to other next generation sequencing platforms. However, a limitation of this method is that it generates only short sequences of DNA, which presents challenges to mapping its reads to a reference genome. After purchasing Complete Genomics, the Beijing Genomics Institute (BGI) refined DNA nanoball sequencing to sequence nucleotide samples on their own platform.
 W
WA DNA sequencer is a scientific instrument used to automate the DNA sequencing process. Given a sample of DNA, a DNA sequencer is used to determine the order of the four bases: G (guanine), C (cytosine), A (adenine) and T (thymine). This is then reported as a text string, called a read. Some DNA sequencers can be also considered optical instruments as they analyze light signals originating from fluorochromes attached to nucleotides.
 W
WEnd-sequence profiling (ESP) is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations.
 W
WExome sequencing, also known as whole exome sequencing (WES), is a genomic technique for sequencing all of the protein-coding regions of genes in a genome. It consists of two steps: the first step is to select only the subset of DNA that encodes proteins. These regions are known as exons – humans have about 180,000 exons, constituting about 1% of the human genome, or approximately 30 million base pairs. The second step is to sequence the exonic DNA using any high-throughput DNA sequencing technology.
 W
WForensic DNA Consultants (Pty) Ltd. (FDC) specialises in the provision of advisory and technical services related to the various aspects of forensic DNA profiling and analysis and their related laboratory, legal and administrative processes.
 W
WJumping libraries or junction-fragment libraries are collections of genomic DNA fragments generated by chromosome jumping. These libraries allow us to analyze large areas of the genome and overcome distance limitations in common cloning techniques. A jumping library clone is composed of two stretches of DNA that are usually located many kilobases away from each other. The stretch of DNA located between these two “ends” is deleted by a series of biochemical manipulations carried out at the start of this cloning technique.
 W
WMaxam–Gilbert sequencing is a method of DNA sequencing developed by Allan Maxam and Walter Gilbert in 1976–1977. This method is based on nucleobase-specific partial chemical modification of DNA and subsequent cleavage of the DNA backbone at sites adjacent to the modified nucleotides.
 W
WThe New York Genome Center (NYGC) is an independent 501(c)(3) nonprofit academic research institution in New York, New York. It serves as a multi-institutional collaborative hub focused on the advancement of genomic science and its application to drive novel biomedical discoveries. NYGC’s areas of focus include the development of computational and experimental genomic methods and disease-focused research to better understand the genetic basis of cancer, neurodegenerative disease, and neuropsychiatric disease. In 2020, the NYGC also has directed its expertise to COVID-19 genomics research.
 W
WOligotyping is the process of correcting DNA sequence measured during the process of DNA sequencing based on frequency data of related sequences across related samples.
 W
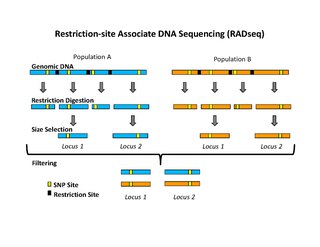
WRestriction site associated DNA (RAD) markers are a type of genetic marker which are useful for association mapping, QTL-mapping, population genetics, ecological genetics and evolution. The use of RAD markers for genetic mapping is often called RAD mapping. An important aspect of RAD markers and mapping is the process of isolating RAD tags, which are the DNA sequences that immediately flank each instance of a particular restriction site of a restriction enzyme throughout the genome. Once RAD tags have been isolated, they can be used to identify and genotype DNA sequence polymorphisms mainly in form of single nucleotide polymorphisms (SNPs). Polymorphisms that are identified and genotyped by isolating and analyzing RAD tags are referred to as RAD markers.
 W
WSingle cell epigenomics is the study of epigenomics in individual cells by single cell sequencing. Since 2013, methods have been created including whole-genome single-cell bisulfite sequencing to measure DNA methylation, whole-genome ChIP-sequencing to measure histone modifications, whole-genome ATAC-seq to measure chromatin accessibility and chromosome conformation capture.
 W
WTrajectory inference or pseudotemporal ordering is a computational technique used in single-cell transcriptomics to determine the pattern of a dynamic process experienced by cells and then arrange cells based on their progression through the process. Single-cell protocols have much higher levels of noise than bulk RNA-seq, so a common step in a single-cell transcriptomics workflow is the clustering of cells into subgroups. Clustering can contend with this inherent variation by combining the signal from many cells, while allowing for the identification of cell types. However, some differences in gene expression between cells are the result of dynamic processes such as the cell cycle, cell differentiation, or response to an external stimuli. Trajectory inference seeks to characterize such differences by placing cells along a continuous path that represents the evolution of the process rather than dividing cells into discrete clusters. In some methods this is done by projecting cells onto an axis called pseudotime which represents the progression through the process.
 W
WTransmission electron microscopy DNA sequencing is a single-molecule sequencing technology that uses transmission electron microscopy techniques. The method was conceived and developed in the 1960s and 70s, but lost favor when the extent of damage to the sample was recognized.
 W
WThe Wellcome Genome Campus is a scientific research campus built in the grounds of Hinxton Hall, Hinxton in Cambridgeshire, England.