 W
WNucleotidyltransferases are transferase enzymes of phosphorus-containing groups, e.g., substituents of nucleotidylic acids or simply nucleoside monophosphates. The general reaction of transferring a nucleoside monophosphate moiety from A to B, can be written as:A-P-N + B A + B-P-N
 W
WIn enzymology, a 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase is an enzyme that catalyzes the chemical reaction:2-C-methyl-D-erythritol 4-phosphate + CTP diphosphate + 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol
 W
WIn enzymology, an adenosylcobinamide-phosphate guanylyltransferase is an enzyme that catalyzes the chemical reactionGTP + adenosylcobinamide phosphate diphosphate + adenosylcobinamide-GDP
 W
WPhosphatidate cytidylyltransferase 1 is an enzyme that in humans is encoded by the CDS1 gene.
 W
WPhosphatidate cytidylyltransferase 2 is an enzyme that in humans is encoded by the CDS2 gene.
 W
WDiadenylate cyclase EC 2.7.7.85, DNA integrity scanning protein DisA is a DNA binding protein participates in a DNA-damage check-point. DisA forms globular foci that rapidly scan along the chromosomes searching for lesions. Catalytic activity2 ATP 2 diphosphate + cyclic di-3',5'-adenylate.
 W
WIn enzymology, diguanylate cyclase, also known as diguanylate kinase, is an enzyme that catalyzes the chemical reaction:
 W
WA DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reactiondeoxynucleoside triphosphate + DNAn ⇌ pyrophosphate + DNAn+1.
 W
WDNA polymerase alpha also known as Pol α is an enzyme complex found in eukaryotes that is involved in initiation of DNA replication. The DNA polymerase alpha complex consists of 4 subunits: POLA1, POLA2, PRIM1, and PRIM2.
 W
WDNA polymerase delta (DNA Pol δ) is an enzyme complex found in eukaryotes that is involved in DNA replication and repair. The DNA polymerase delta complex consists of 4 subunits: POLD1, POLD2, POLD3, and POLD4. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis. It exhibits increased processivity when interacting with the proliferating cell nuclear antigen (PCNA). As well, the multisubunit protein replication factor C, through its role as the clamp loader for PCNA is important for DNA Pol δ function.
 W
WDNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
 W
WDNA polymerase II is a prokaryotic DNA-Dependent DNA polymerase encoded by the PolB gene.
 W
WDNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg and Malcolm Gefter in 1970. The complex has high processivity and, specifically referring to the replication of the E.coli genome, works in conjunction with four other DNA polymerases. Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork.
 W
WGalactose-1-phosphate uridylyltransferase is an enzyme responsible for converting ingested galactose to glucose.
 W
WIn enzymology, a glucose-1-phosphate adenylyltransferase is an enzyme that catalyzes the chemical reactionATP + alpha-D-glucose 1-phosphate diphosphate + ADP-glucose
 W
WIn enzymology, a glucose-1-phosphate thymidylyltransferase is an enzyme that catalyzes the chemical reactiondTTP + alpha-D-glucose 1-phosphate diphosphate + dTDP-glucose
 W
WIn enzymology, a N-acylneuraminate cytidylyltransferase is an enzyme that catalyzes the chemical reactionCTP + N-acylneuraminate diphosphate + CMP-N-acylneuraminate
 W
WIn enzymology, nicotinamide-nucleotide adenylyltransferase (NMNAT) (EC 2.7.7.1) are enzymes that catalyzes the chemical reactionATP + nicotinamide mononucleotide diphosphate + NAD+
 W
WIn enzymology, a pantetheine-phosphate adenylyltransferase is an enzyme that catalyzes the chemical reactionATP + 4'-Phosphopantetheine diphosphate + 3'-dephospho-CoA
 W
WDNA-directed RNA polymerase, mitochondrial is an enzyme that in humans is encoded by the POLRMT gene.
 W
WA polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.
 W
WIn enzymology, a polynucleotide adenylyltransferase is an enzyme that catalyzes the chemical reactionATP + RNA-3'OH pyrophosphate + RNApA-3'OH
 W
WPolynucleotide Phosphorylase (PNPase) is a bifunctional enzyme with a phosphorolytic 3' to 5' exoribonuclease activity and a 3'-terminal oligonucleotide polymerase activity. That is, it dismantles the RNA chain starting at the 3' end and working toward the 5' end. It also synthesizes long, highly heteropolymeric tails in vivo. It accounts for all of the observed residual polyadenylation in strains of Escherichia coli missing the normal polyadenylation enzyme. Discovered by Marianne Grunberg-Manago working in Severo Ochoa's lab in 1955, the RNA-polymerization activity of PNPase was initially believed to be responsible for DNA-dependent synthesis of messenger RNA, a notion that got disproved by the late 1950s.
 W
WA reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.
 W
WIn molecular biology, RNA polymerase, is an enzyme that synthesizes RNA from a DNA template.
 W
WRNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.
 W
WRNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
 W
WRNase PH is a tRNA nucleotidyltransferase, present in archaea and bacteria, that is involved in tRNA processing. Contrary to hydrolytic enzymes, it is a phosphorolytic enzyme, meaning that it uses inorganic phosphate as a reactant to cleave nucleotide-nucleotide bonds, releasing diphosphate nucleotides. The active structure of the proteins is a homohexameric complex, consisting of three ribonuclease (RNase) PH dimers. RNase PH has homologues in many other organisms, which are referred to as RNase PH-like proteins. The part of another larger protein with a domain that is very similar to RNase PH is called an RNase PH domain (RPD).
 W
WIn enzymology, a sulfate adenylyltransferase is an enzyme that catalyzes the chemical reactionATP + sulfate pyrophosphate + adenylyl sulfate
 W
WT7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
 W
WTelomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most eukaryotes. Telomeres protect the end of the chromosome from DNA damage or from fusion with neighbouring chromosomes. The fruit fly Drosophila melanogaster lacks telomerase, but instead uses retrotransposons to maintain telomeres.
 W
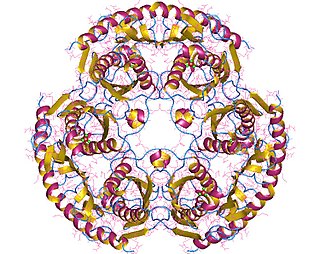
WTerminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphoma cells. TdT adds N-nucleotides to the V, D, and J exons of the TCR and BCR genes during antibody gene recombination, enabling the phenomenon of junctional diversity. In humans, terminal transferase is encoded by the DNTT gene. As a member of the X family of DNA polymerase enzymes, it works in conjunction with polymerase λ and polymerase μ, both of which belong to the same X family of polymerase enzymes. The diversity introduced by TdT has played an important role in the evolution of the vertebrate immune system, significantly increasing the variety of antigen receptors that a cell is equipped with to fight pathogens. Studies using TdT knockout mice have found drastic reductions (10-fold) in T-cell receptor (TCR) diversity compared with that of normal, or wild-type, systems. The greater diversity of TCRs that an organism is equipped with leads to greater resistance to infection. Although TdT was one of the first DNA polymerases identified in mammals in 1960, it remains one of the least understood of all DNA polymerases. In 2016–18, TdT was discovered to demonstrate in trans template dependant behaviour in addition to its more broadly known template independent behaviour
 W
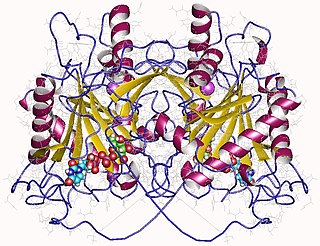
WIn enzymology, a tRNA nucleotidyltransferase is an enzyme that catalyzes the chemical reactiontRNAn+1 + phosphate tRNAn + a nucleoside diphosphate
 W
WIn enzymology, an UDP-glucose—hexose-1-phosphate uridylyltransferase is an enzyme that catalyzes the chemical reactionUDP-glucose + alpha-D-galactose 1-phosphate alpha-D-glucose 1-phosphate + UDP-galactose
 W
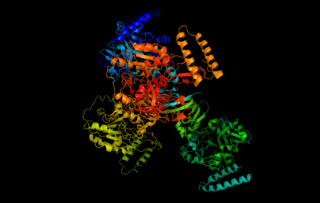
WIn enzymology, an UDP-N-acetylglucosamine diphosphorylase is an enzyme that catalyzes the chemical reactionUTP + N-acetyl-alpha-D-glucosamine 1-phosphate diphosphate + UDP-N-acetyl-D-glucosamine
 W
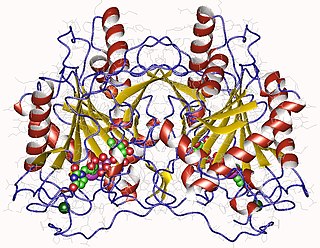
WUTP—glucose-1-phosphate uridylyltransferase also known as glucose-1-phosphate uridylyltransferase is an enzyme involved in carbohydrate metabolism. It synthesizes UDP-glucose from glucose-1-phosphate and UTP; i.e.,glucose-1-phosphate + UTP UDP-glucose + pyrophosphate
 W
WIn enzymology, an UTP—hexose-1-phosphate uridylyltransferase is an enzyme that catalyzes the chemical reactionUTP + alpha-D-galactose 1-phosphate diphosphate + UDP-galactose